-
 FagoVal: Selective filtering of raster values
FagoVal: Selective filtering of raster values
Direct access to online help: FagoVal
Access the application from the menu:
"Tools | Image classification | Selective filter"
"Tools | Image processing | Selective filter"
"Tools | Cartographic generalization | Raster | Selective generalization"
Presentation and options
This application takes a raster and substitutes the cells (pixels) which have a value equal to a previously selected value (value to substitute) with the value of the majority of the cells surrounding that cell (mode) or the average of the neighboring values. The neighborhood is defined by the convolution window, which is square and has an odd number of pixels. The file to be processed is a raster in any format (byte, short integer, unsigned integer o real), compressed or uncompressed. The result of the process will be another raster in the same format.
Conceptually, this process can have a number of uses:
- In image processing, as a selective filter that eliminates certain values.
- After classifying a remote sensing image, for substituting the unclassified pixels (typically zeros) with the most frequent value in each neighborhood.
- In cartographic and GIS generalization, and also after image classification, to assign certain values (typically the result of selecting a small group of cells) to the most frequent values (categories) in the neighborhood. This process is geographically much better than the traditional mode filters, which are too indiscriminate and change the shapes of polygons excessively.
The name of the application, FagoVal, is due to the similarity between the process carried out and the process of biological phagocytosis by which a larger cell (in this case the convolution matrix) eliminates other elements (in this case the selected values) by surrounding them and ingesting them into itself.
When the area of the values to be substituted is bigger than the convolution window, some interior values will remain after the first pass of the process and in this case the application asks the users if they wish to perform another iteration of the substitution process. In order to avoid interrupting the application and continue the iteration until there are no more values to process or until there are only isolated values, there is the optional parameter /ITERAR_AUTO.
It may also be the case that a value is not phagocytosed because the result of the substitution is itself according to the convolution calculation (for example, in a substitution by average or also in a case by median with tiebreaker by average of two central values).
To avoid final isolated values, that appear when a value to be processed is surrounded by NoData values, and to assign them the value NoData, there is the optional parameter /AILLAT_NODATA. Prudence is required when choosing the size of the convolution window; whilst a bigger window will reduce the number of iterations, it may also degrade the image by applying a substitute value from excessively distant cells.
When replacing the values to be processed by the mean or modal value of the convolution window, NoData values are never considered, which can result in the calculation being made on a very limited sample. In this case, if the number of valid pixels within the convolution window is inferior to half the number of cells in the window, it is possible to mark the pixel that is being processed with a NoData value if the /NODATA_MAJORITARI parameter is used. Alternatively, accept the calculation made with the available sample.
In quantile calculations, such as the median, it is possible to indicate, with the modifier /MEDIANA_EMPAT=, the type of tiebreaker to use for its calculation when the position of the quantile is between two values of the series. For more information, see general syntax.

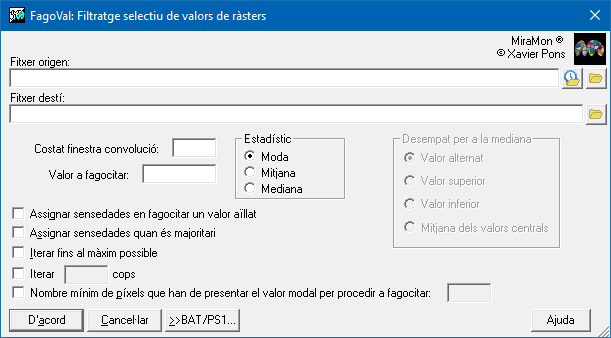
Dialog box of the application

Syntax
Syntax:
- FagoVal OriginFile OutputFile Convol_Win Value_phago Statistic [/AILLAT_NODATA] [/NODATA_MAJORITARI] [/ITERAR_AUTO] [/NO_ITERAR] [/NODATA] [/NMIN] [/MEDIANA_EMPAT]
Parameters:
- OriginFile
(Raster input file -
Input parameter): Is the raster file with values to be substituted.
- OutputFile
(Raster output file -
Output parameter): The resulting raster file after the substitution process.
- Convol_Win
(Convolution window -
Input parameter): The number of pixels in the side of the convolution window (an odd number), typically 3 or 5.
- Value_phago
(Value to phagocytize -
Input parameter): Represents the value to be substituted by the statistical calculation performed within its convolution window. It can be "NoData" to process the NoData value.
- Statistic
(Statistic -
Input parameter): The statistic to use in the calculation: M (mode), m (mean) or d (median).
Modifiers:
/AILLAT_NODATA
(Isolated as NoData)
The value to be processed will be marked as background value (NoData) when it be completely surrounded by NoData values, that is, when the convolution window contains the pixel being processed in the form of a valid value and the rest of the window with NoData values. If this parameter is omitted, the value surrounded by NoData will never be processed, except if the process is repeated with a bigger convolution window. (Input parameter) /NODATA_MAJORITARI
(Assign to NoData)
The pixel to be processed will be assigned to NoData value when the sample of valid values within the convolution window is inferior to half the number of cells in it. If this option is omitted then calculations will be performed in all cases when there is at least a single valid value. In this case, NoData values will simply be ignored. (Input parameter) /ITERAR_AUTO
(Indefinite iterations)
Perform an indefinite number of iterations in order to keep reducing the number of pixels to be processed. The process will end when there are no more values to be processed or there are only isolated values which could not be processed in further iterations (this will only happen in the absence of the /AILLAT_NODATA parameter). If this parameter is not present, the user is prompted after each iteration in order to know whether to continue the process or stop it. (Input parameter) /NO_ITERAR
(One iteration)
Apply only one iteration step even if there are pixels left to process. (Input parameter) /NODATA
(Process NoData values)
Process NoData values (Input parameter) /NMIN=
(Minimum number of pixels presenting the modal value to proceed phagocytizing)
Minimum number of pixels presenting the modal value to proceed phagocytizing. It can only be used in the case of mode and must be between 1 and (side of the window)^2 - 1. (Input parameter) /MEDIANA_EMPAT=
(Decision for quantiles)
If the calculation of a quantile (such as the median, a quartile or a percentile) has been requested, it indicates the tiebreaking criterion to be used for its calculation. To learn more about the values of this parameter, please consult general syntax document. (Input parameter)